A case study in happy extremophiles
The touchy science behind achieving a pure, thriving culture of methanotrophs
If asked to describe your ideal environment, the odds are you wouldn’t opt for somewhere exceedingly salty, with an acidic pH or a dense supply of methane. However, some organisms (with fewer cells and vastly different standards than you and me) would say that sounds just about perfect.
Researchers recently visited Sanford Underground Research Facility (Sanford Lab) to collect samples of organisms that prefer the damp, dark environment of the deep subsurface. Now, they are trying to replicate those seemingly abysmal conditions back in their laboratory. By providing the perfect conditions, researchers can selectively grow the bacteria they want to study.
This research is part of the National Science Foundation’s (NSF) BuG ReMeDee project (Building Genome-to-Phenome Infrastructure for Regulating Methane in Deep and Extreme Environments). This collaborative group of researchers from three universities is seeking to understand curious life forms called methanotrophs—organisms that survive by consuming methane.
“Much of the general public looks at bacteria like germs, like something harmful,” said Christopher Abin, postdoctoral researcher at the University of Oklahoma. “But what we see is that the vast majority of bacteria are incredibly important—without them, the earth wouldn't really function properly. In fact, life on earth would cease to exist without bacteria.”
In the effort to understand and utilize the creatures that feast on a greenhouse gas more potent than carbon dioxide, each collaborating university has their niche.
At South Dakota School of Mines & Technology (SD Mines), principal investigator Rajesh Sani’s team focuses on genetically engineering and improving methane-consuming microbes to create useable products and materials, such as biofuels, biodegradable plastics or electricity. At Montana State University (MSU), Robin Gerlach’s team is developing models that show how microbes consume methane and create energy. This helps scientists better understand how methane generated under such places as Yellowstone National Park and other geothermal environments and fossil fuel beds impacts our climate.
But before models can be made and genes engineered, researchers need a solid understanding of how these organisms function. To study them in detail, researchers from the University of Oklahoma (OU) and under the lead of Lee Krumholz, collect and cultivate samples, isolating pure cultures of methanotrophs in the lab. There’s just one small setback: the organisms of interest come from some of our planet’s most extreme environments—environments that are quite difficult to replicate in a laboratory.
As the microbe-hunters of the group, OU researchers go to various extreme environments—hot springs, lakes ten times saltier than the ocean, sulfur springs with no measurable oxygen content and locations in the deep subsurface, miles below the earth—in search of methanotrophs.
“We don’t fully understand the flux of methane in these extreme environments,” said Abin. “These locations could be either a sink or a source of methane to the atmosphere. Little research has been devoted to understanding the microbes that inhabit these areas, so any samples we collect can be novel.”
At Sanford Lab, researchers traveled deep underground to collect samples from biofilms and groundwater from boreholes on the 4850 Level and sediments from the 1700 Level.
“We also collected a sample from an exotic fungus growing on a wooden beam,” Abin said. “You don't really know going in what you're going to find, so you sample everything you think might be interesting. You might discover something really cool when you analyze it back in the lab.”
During each excursion, the team takes two sets of samples. The first is dedicated to a DNA roll-call that identifies the hundreds—perhaps thousands—of species naturally present in that environment. The second set is dedicated to an advanced cultivation process in the lab, where researchers try to single out one or a few specific species through a process called enrichment.
“In the lab, we put our samples in bottles that can be sealed completely then add concentrations of gases like methane and oxygen at precise concentrations. As the methanotrophs consume methane, the concentration slowly decreases,” explained Abin.
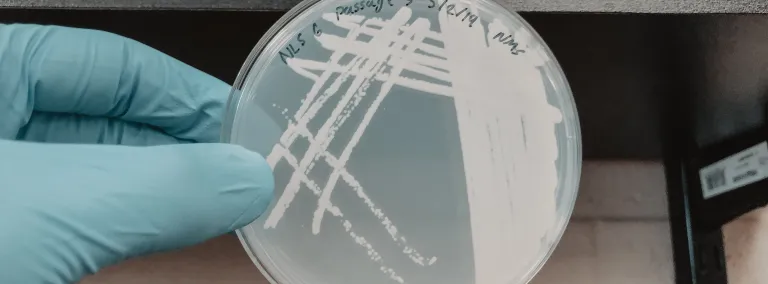
“Once it is mostly depleted, we dilute the cultures to get rid of the background microbes we don't want, achieving a higher proportion of just the methanotrophs," said Abin. "We take a small amount of that liquid and place it onto a petri plate containing a semisolid material called agar to provide a substrate for the bacteria to grow on. As the bacteria grow, they produce visible colonies that we can purify further through a process called streaking.”
At the end of the streaking process, researchers hope to isolate the single species from the multitudes present. Sometimes, however, organisms resist, preferring a more social environment.
“Species don't grow in pure cultures in their natural environment,” said Rosie Moon-Escamilla, a graduate student at OU. “When two organisms exist together and provide benefits to each other, it’s difficult to make them survive without each other.”
If the methanotroph is growing in co-culture with another organism that is providing some sort of benefit to them, such as removing toxic substances or suppling a certain vitamin, the isolation process can get complicated.
“You do a lot of work to get the organism isolated, always knowing in the back of your mind that they are happier in co-culture,” said Moon-Escamilla. “Sometimes, it may not be impractical to isolate them into distinct pure cultures, it may be impossible.”
At the end of multiple rounds of streaking, if researchers have achieved a pure culture, they can begin to characterize them—What temperatures do they enjoy? Which solidities do they fancy?—to better understand the microbial preferences.
“The challenge of microbiology cultivation in general is how to replicate the environment you sample from,” said Christopher Garner, an OU graduate student. “We estimate that 90 percent of all microorganisms out there haven't been cultivated in the lab, because it's just something that's really hard to do. When your samples are from an extreme environment, that adds additional challenges that makes it more difficult to cultivate.”
Collected from vastly differing locations, the physiology of these organisms varies—each suited to its own extreme environment—and each must be assessed and studied individually. The binding commonality, however, is that these organisms use methane as their energy source and have enormous potential in bioengineering applications.
“We've done a lot of work with media manipulation,” Moon-Escamilla said. “If you tailor the media to the specific location, being mindful of the salt and pH levels or different minerals present at each collection site, you have a better chance of increasing the number of microbes that will grow in the lab.”
Much of the work involves experimentation, testing the conditions and letting the organism’s response inform the process.
“There is immense value in traditional microbiology work—cultivating microbes from the environment and learning about their metabolisms,” Garner said. “We've only begun to understand really how many different kinds of microbes there are out there.”
“The overarching goals of the BuG ReMeDEE consortium are to investigate methane cycling in deep and extreme environments and develop new biological routes for converting methane into value-added products,” said principal investigator Rajesh Sani. “Using ‘genome-to-phenome’ approaches, the consortium and will address critical regional, national and global issues of methane cycling, global warming, renewable energy and carbon neutrality.”
“This collaboration will allow our groups to synergistically solve problems that could not be dealt with alone. I feel strongly that our work on isolating and better understanding methanotrophs at SURF and other locations will allow us to better understand the fate of methane and its role as a greenhouse gas,” said Lee Krumholz, who leads the work being done at OU.